RNA-KG
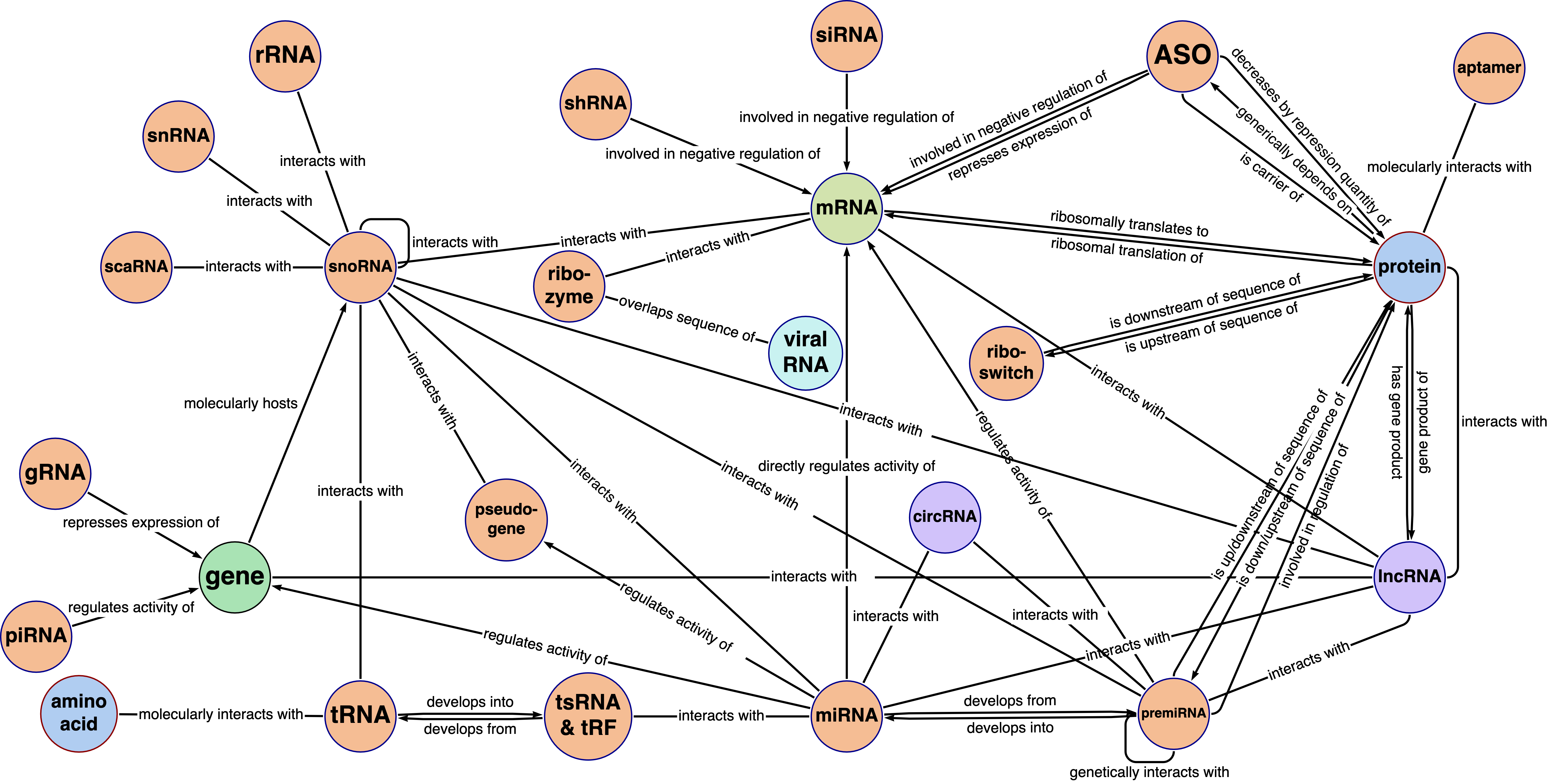
RNA-KG is an ontology-based knowledge graph which encompasses biological knowledge about RNA molecules gathered from more than 60 public data sources, integrating functional relationships with genes, proteins, and chemicals and ontologically grounded biomedical concepts. RNA-KG is constantly maintained and updated with new experimental data. RNA-KG releases are updated on this webpage. More details can be found in our paper.
- Public SPARQL endpoint is available and maintained at the following link. The list of RNA-KG nodes is stored in: nodes.csv; the list of RNA-KG edges is stored in: edges.csv.
- RNA-KG can be built from scratch by following the tutorials on the GitHub page (specifically, notebooks contains Python notebooks to build RNA-KG current release). Data for reproducing RNA-KG contruction and experiments are available at doi/10.5281/zenodo.10078876.
- ⚠️Experimental⚠️ Public Neo4j endpoint is available and maintained at the following link (usr: anacleto; pwd: anacleto). The list of RNA-KG nodes including properties is stored in: nodes_with_properties.csv; the list of RNA-KG edges including properties is stored in: edges_with_properties.csv.
- At the following link, we provide examples and tutorials on how to query RNA-KG and extract relevant subgraphs of interest, generate from scratch new RNA-KG views by means of the PheKnowLator ecosystem, and how to import KGs in the GRAPE library to infer new knowledge from RNA-KG using different kinds of embedding.
While querying RNA-KG using SPARQL is straightforward and takes a few seconds to extract an RNA-KG subgraph of interest, using PheKnowLator for generating a new view ensures to obtain a subgraph of RNA-KG that includes selected ontology relationships.
-
At the following link, we provide views already generated by our team, whose specification is reported for each view in a README.txt file. These views are thought to be used in combination with graph-oriented ML techniques for edge and node type labeling and heterogeneous/homogeneous link predictions tasks. For each view, we also provide the correspondent metagraph (in pdf and xlsx formats) and two pickle dictionaries files specifying nodes and edges to facilitate the import in the GRAPE environment.
-
Finally, at the following link, we provide tabular files for each couple of interactors within RNA-KG. These files can be used for obtaining RNA-KG views to be imported in graph-oriented ML tools/environments different from GRAPE.
Don't hesitate to contact us, especially if you believe a new data source should be integrated into RNA-KG. To get in touch with us, please create an issue on GitHub or send us an email 📩.
Citing RNA-KG
Please cite the following paper if RNA-KG was useful for your research:
@article{Cavalleri2024rnakg,
title="An ontology-based knowledge graph for representing interactions involving RNA molecules",
author="Emanuele Cavalleri and Alberto Cabri and Mauricio Soto-Gomez and Sara Bonfitto and Paolo Perlasca and Jessica Gliozzo and Tiffany J. Callahan and Justin Reese and Peter N Robinson and Elena Casiraghi and Giorgio Valentini and Marco Mesiti",
year="2024",
journal="Sci. Data",
publisher="Springer Science and Business Media LLC",
volume=11,
number=1,
pages="906",
month=aug,
year=2024,
copyright="https://creativecommons.org/licenses/by-nc-nd/4.0",
language="en"
}
